
Davis Lab's work and impact.
The Davis Lab works at the nexus of immunology, epigenetics, smooth muscle cell biology and vascular biology. Using a complement of biochemical assays, in vivo animal models, and biobank of vascular tissue specimens, we employ a range of standard and advanced techniques to investigate pathological pathways of interest.
Monocyte-Macrophage Plasticity & Epigenetic Modification
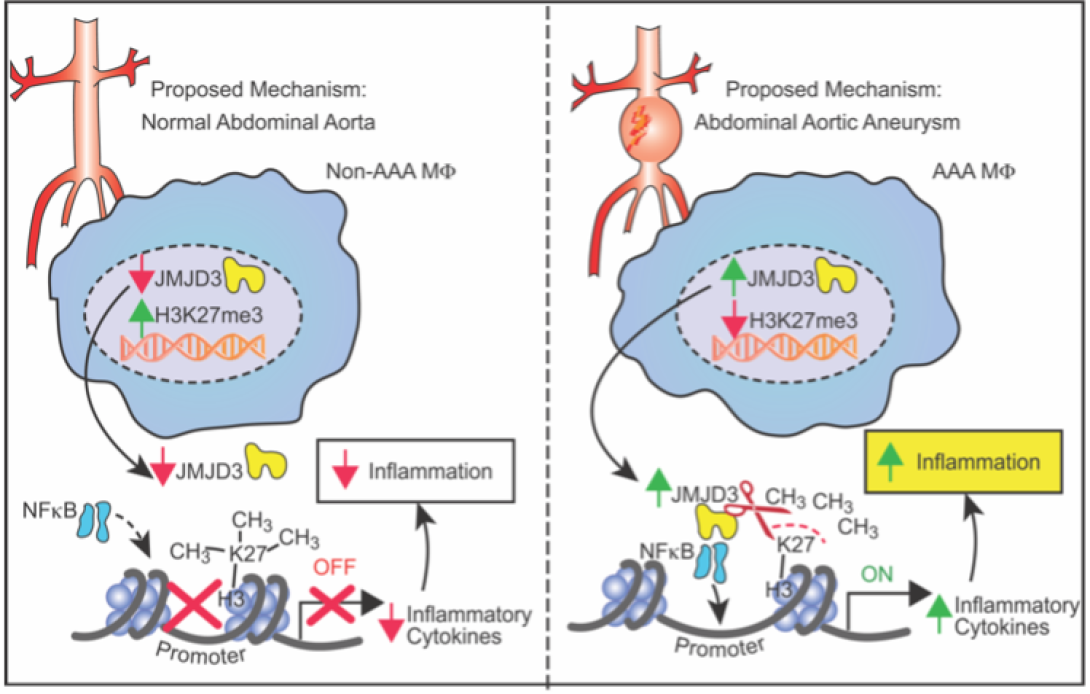
The laboratory focuses on investigating the regulation of monocyte/macrophage polarization during aortic aneurysm development. We were the first to demonstrate that the histone demethylase, JMJD3, was significantly increased in human and murine AAA disease and resulted in epigenetic activation of multiple proinflammatory pathways (PMID: 33779682). Further, pharmacological and genetic inhibition of JMJD3 prevented aortic dilation and rupture in our murine models. Ongoing projects now include investigator lead clinical trials on JMJD3 inhibition as well as bench-top studies evaluating the implications of other epigenetic enzymes in immune cell infiltration. Understanding the triggers that prime these cells toward a proinflammatory state that drive aortic wall degradation is an important research question with tremendous clinical implications.
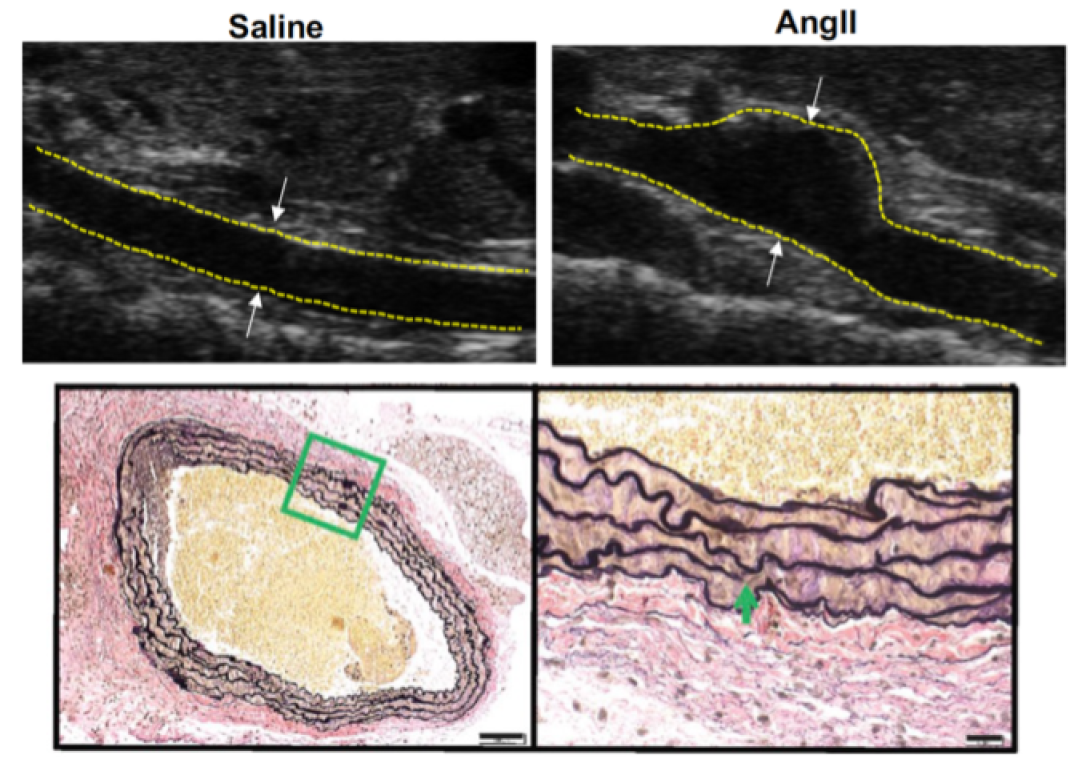
A pathological hallmark of AAA development is smooth muscle cell apoptosis leading to weakening of the aortic wall and dilation. We have identified that mitochondrial and endoplasmic reticulum stress responses are activated in aortic aneurysmal disease resulting in vascular smooth muscle cell inflammation, phenotypic switching, and ultimate apoptosis. We utilize murine genetic models, high-fidelity biochemical assays, and state-of-the-art molecular techniques to help identify potential drug targets that can ultimately impact the clinical progression of AAAs.
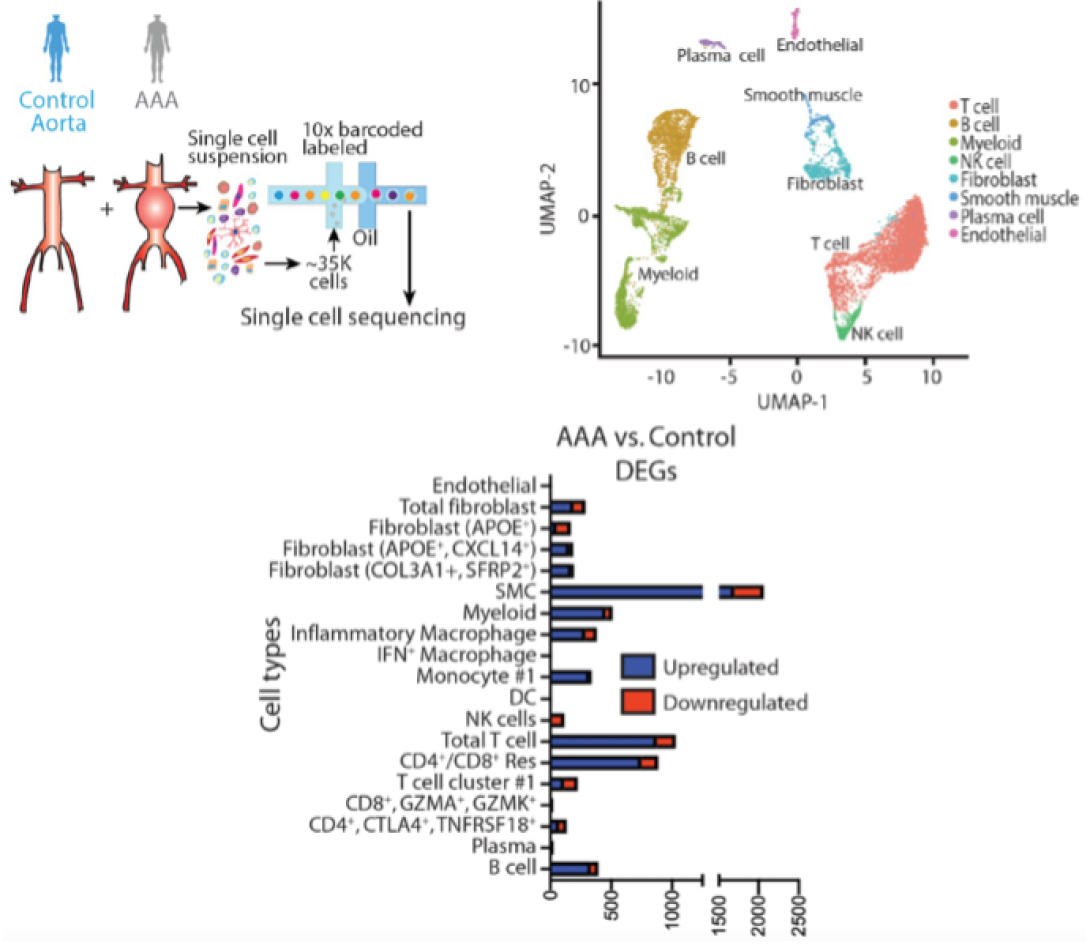
In a quest to improve the understanding of pathological pathways that drive AAA development, our laboratory was the first to conduct a detailed single-cell RNA sequencing of the human abdominal aorta (PMID: 35762613 and 33779682). This demonstrated multiple pathologic myeloid and T cell subsets within the aortic wall as well as increased ligand-receptor interactions between myeloid and smooth muscle cells (mainly CCL2/CCR2, JAG/Notch2, TGFB1/TGFB1R and CXCL12/CXCR4) that appear to impede the normal smooth muscle cell function. Further, we integrated our single cell data with prior GWAS aortic aneurysm data to demonstrate the potential pathological role of SORT1 in vascular smooth muscle cells. Currently we are exploring spatial sequencing and imaging mass cytometry to provide spatial context for different cellular and gene expression within the aortic wall.
Clinical research conducted by investigators in our laboratory has changed how we practice in the clinic and helped fine-tune care. Using large statewide databases, we have sought to improve the care of patients with vascular disease. Aortic rupture is the most devastating consequence of an aortic aneurysm and timely intervention is warranted. As such, we sought to optimize the “door-to-intervention” time for the management of aortic aneurysm rupture designing the optimal window for surgical repair (PMID 31201978).
Further, for vascular surgery patients, myocardial infarction and surgical site infections are two of the most common and most morbid postoperative complications following abdominal aortic aneurysm repair (PMID 28527931 and 32473343). We identified preoperative and intraoperative risk factors for postoperative myocardial infarctions and surgical site infection and are working to design clinical care algorithms to decrease these postoperative risks. The long-term goal is to improve outcomes for vascular surgery patients by mitigating the risk factors for complications.
- National Institutes of Health
- American Heart Association
- Society of University Surgeons
- Vascular and Endovascular Surgery Society
- American Surgical Association
- Lefkofsky Scholars Research Fund