
All RNA and DNA entering the AGC undergoes quality control. Information regarding assays employed, interpreting results, and QC only requirements.
All nucleic acids are subject to quality control in order to identify possible low-performing samples before they are used in protocols.
If samples do not meet our requirements for input amounts and/or concentration, you will receive an iLab notification detailing the QC findings and requesting your review of the data. Your project will be put on hold until permission to proceed is received. Via the comment section in your iLab request, you may request samples be omitted, ask to submit replacement samples, or choose to proceed with samples that have lower than required amounts and/or poor integrity.
Please note that the Advanced Genomics Core can almost always make a NGS library out of low and/or poor quality input amounts. These libraries will yield standard TapeStation traces and a nM concentration as measured by Kapa qPCR. If we see extremely low yields, excess adapters, or unusual profiles, you will be notified via iLab to determine if the libraries should be pulled from sequencing or subjected to additional processing (additional charge may apply). It is important to note that libraries generated from low and/or poor quality samples can exhibit lower complexity and an increased duplication rate which will be reflected in the sequencing data.
Nucleic acids isolated from FFPE samples will be poor quality. The core has modified protocols designed to maximize the chance of success, but you should be prepared for samples to drop out during processing. The more input you can provide for FFPE samples, the better the chance of success.
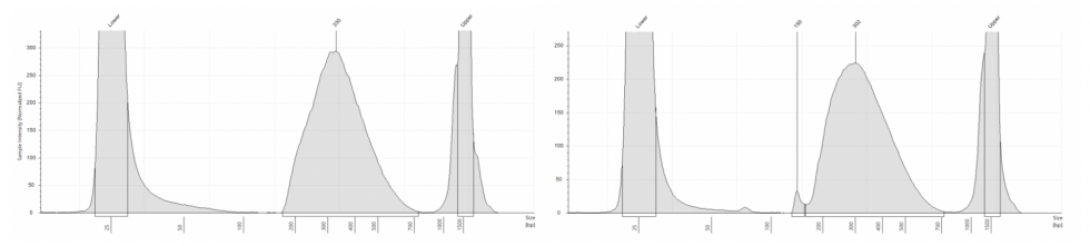
AGC DNA Quality Control includes:
- Quantification using Qubit fluorescence-based assays
- Visualization of DNA size on Agilent TapeStation
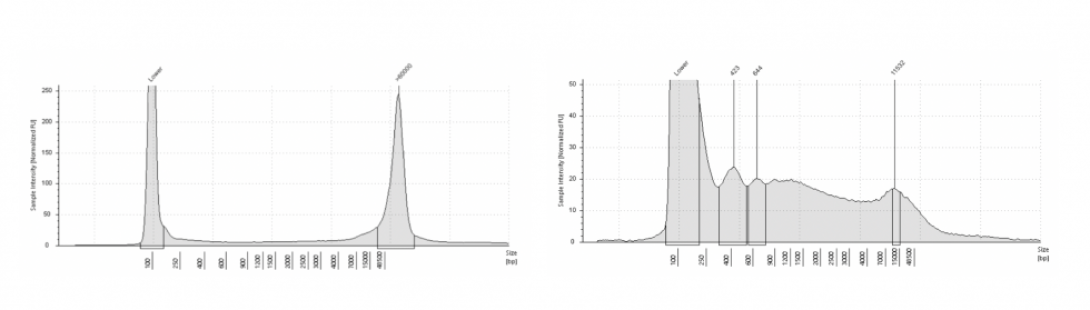
AGC RNA Quality Control includes:
- Quantification via Qubit fluorescence-based assays
- Visualization of RNA size on Agilent TapeStation
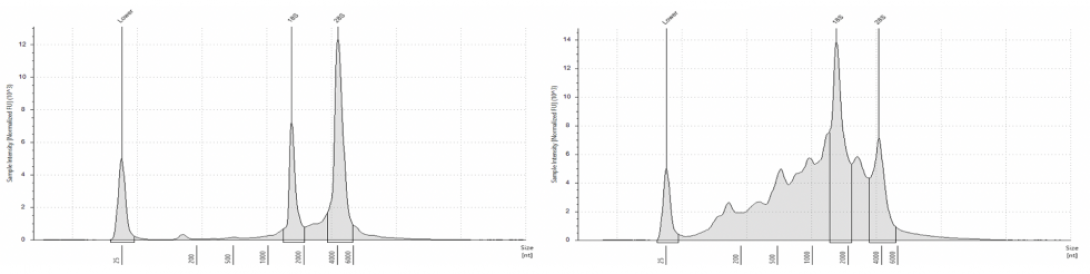
AGC Library Quality Control includes:
- Quantification via Qubit
- Visualization of DNA size on Agilent TapeStation
- Kapa real-time PCR to determine final concentration for sequencing
The Advanced Genomics Core accepts samples for quality control assessment only. Submissions go through iLab by initiating a Nucleic-Acid Quality Control request. For either DNA or RNA QC an aliquot of 5 µl of sample is required. DO NOT submit stock samples for analysis! Samples submitted for analysis will not be returned to researchers and will be discarded after 2 weeks. All samples are first subjected to Qubit Assay to determine concentration and inform selection of appropriate quality assay. All quality assessments samples will be done on the TapeStation, unless otherwise requested. Fees are charged per sample; you may be charged for additional samples if you are not running a full column (TapeStation) or cartridge (Bioanalyzer) and we do not have enough other samples to fill in the run.
| Quality Check Service Options | Functional Range |
| Bioanalyzer DNA 1000 chip | 100 pg/uL to 50 ng/uL |
| Bioanalyzer DNA 7500 chip | 500 pg/uL to 50 ng/uL |
| Bioanalyzer DNA 12,000 chip | 500 pg/uL to 50 ng/uL |
| Bioanalyzer DNA High Sensitivity chip (up to 7500 bp) | 5 pg/uL to 500 pg/uL |
| Bioanalyzer RNA Nano chip (up to 6000 nt) | 5 ng/uL to 500 ng/uL |
| Bioanalyzer RNA Pico chip (up to 6000 nt) | 50 pg/uL to 5000 pg/uL |
| Bioanalyzer Small RNA chip (6 to 150 nt) | 50 pg/uL to 5000 pg/uL |
| TapeStation DNA 1000 Tape (35 bp to 1000 bp) | 100 pg/uL to 50 ng/ul |
| TapeStation DNA High Sensitivity 1000 Tape (35 bp to 1000 bp) | 10 pg/uL to 1000 pg/uL |
| TapeStation Genomic DNA Tape (200 bp to > 60,000 bp) | 5 ng/uL to 300 ng/uL |
| TapeStation RNA Tape (100 nt to 6000 nt) | 25 ng/uL to 500 ng/uL |
| TapeStation High Sensitivity RNA Tape (100 nt to 6000 nt) | 1 ng/uL to 25 pg/uL |
| Qubit Broad Range dsDNA Assay | 0.5 ng/uL to 1000 ng/uL |
| Qubit High Sensitivity dsDNA Assay | 10 pg/uL to 50 ng/uL |
| Qubit ssDNA Assay | 50 pg/uL to 200 ng/uL |
| Qubit RNA Assay | 1 ng/uL to 1000 ng/uL |
| Qubit High Sensitivity RNA Assay | 250 pg/uL to 100 ng/uL |
| NanoDrop Spectrophotometer | 10 ng/uL to 2500 ng/uL |
For more information about each service option, refer the manufacturer’s websites for the Bioanalzyer & TapeStation and the Qubit & NanoDrop.
There is notable variability between concentrations quantified spectrophotometrically (e.g. Nanodrop) as compared to fluorometrically (e.g. Qubit). While spectroscopic methods provide adequate estimation at concentrations higher than 10 ng/µl, fluorometric quantifications are more precise at lower concentrations. For this reason, all samples are quantified by Qubit upon entering the AGC.
On an agarose gel, DNA contamination will be visible as a smear of band of fragments considerably larger than the RNA (>10 kb). On Bioanalyzer or Tapestation RNA-chips, DNA will be visible in the size range from 4 to 10 kb.
Nucleic acids and proteins have absorbance maxima at 260 and 280 nm, respectively. The ratio of absorbances at these wavelengths has historically been used as a measure of RNA or DNA “purity”. A ratio of ~1.8 is generally accepted as “pure” for DNA whereas a ratio of ~2.0 is preferred for RNA. Many samples fail to reach these minimum cutoffs. 260/280 ratios will fall below the acceptable limits if the sample concentration is low (<10ng/μl) or if the sample is contaminated by protein or other reagents such as phenol. Although not considered “pure”, these samples often perform well in downstream applications.
University of Michigan
2800 Plymouth Rd.
Ann Arbor, MI 48109-2800